Our research contributes to the mapping of pancreatic ß-cell by the Pancreatic ß-Cell Consortium
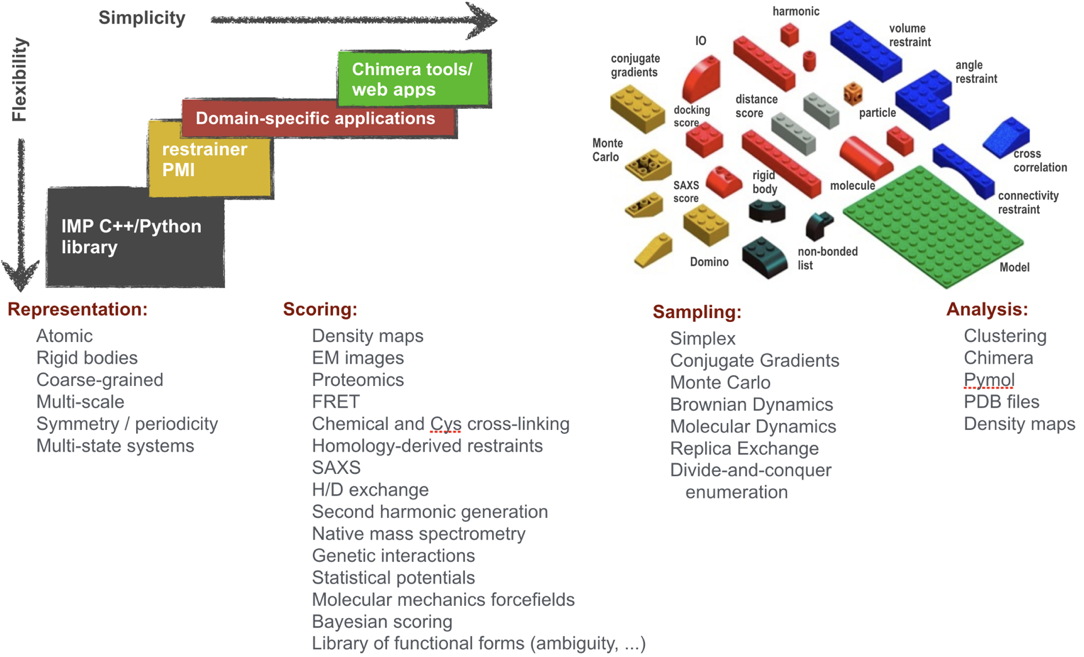
Integrative Modeling Platform
We have been contributing to Integrative Modeling Platform (IMP), an open-source software package that provides programmatic support for constructing and distributing integrative structure modeling protocols. We also demonstrated its use by application to more than 30 biological systems, including the Nuclear Pore Complex (NPC) and 26S proteasome. IMP already allows representing molecules at multiple resolutions, using spatial restraints from many types of data, and searching for solutions by a variety of sampling algorithms. So far, it has been applied mostly to data from electron microscopy (EM), small angle X-ray scattering (SAXS), Förster Resonance Energy Transfer (FRET), cysteine and chemical cross-linking, hydrogen deuterium exchange (HDX), as well as various proteomics methods. IMP is easily extensible to add support for new data sources and algorithms, and is distributed under an open source license. We are building upon this existing foundation, to be able to address a greater range of biological problems and make IMP more generally useful to the scientific community.
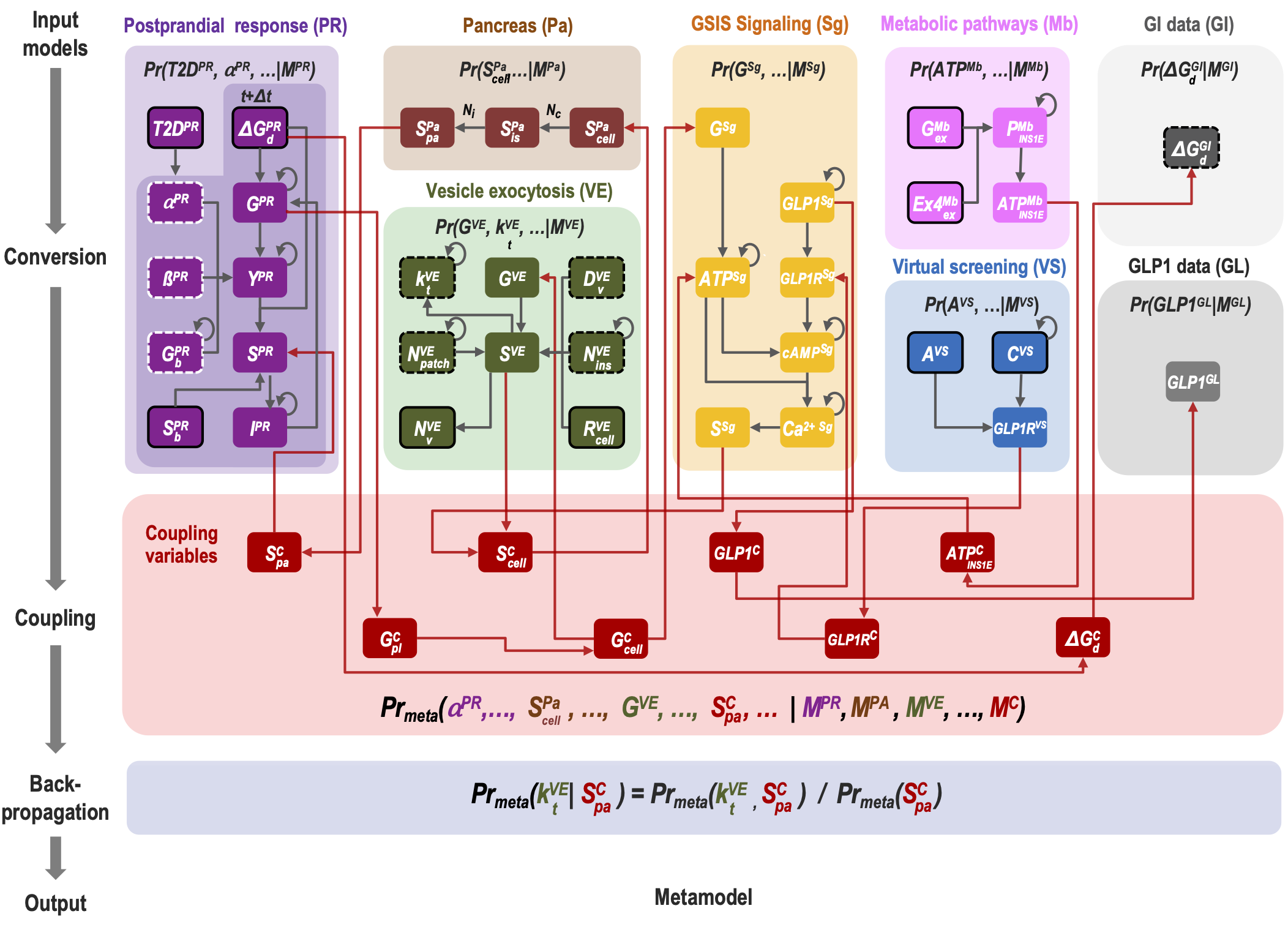
Bayesian Meta-Modeling
Comprehensive modeling of a whole cell requires an integration of vast amounts of information on various aspects of the cell and its parts. To divide and conquer this task, we introduce Bayesian metamodeling, a general approach to modeling complex systems by integrating a collection of heterogeneous input models. Each input model can in principle be based on any type of data and can describe a different aspect of the modeled system using any mathematical representation, scale, and level of granularity. These input models are 1) converted to a standardized statistical representation relying on probabilistic graphical models, 2) coupled by modeling their mutual relations with the physical world, and 3) finally harmonized with respect to each other. To illustrate Bayesian metamodeling, we provide a proof-of-principle metamodel of glucose-stimulated insulin secretion by human pancreatic β-cells. The input models include a coarse-grained spatiotemporal simulation of insulin vesicle trafficking, docking, and exocytosis; a molecular network model of glucose-stimulated insulin secretion signaling; a network model of insulin metabolism; a structural model of glucagon-like peptide-1 receptor activation; a linear model of a pancreatic cell population; and ordinary differential equations for systemic postprandial insulin response. Metamodeling benefits from decentralized computing, while often producing a more accurate, precise, and complete model that contextualizes input models as well as resolves conflicting information. We anticipate Bayesian metamodeling will facilitate collaborative science by providing a framework for sharing expertise, resources, data, and models, as exemplified by the Pancreatic β-Cell Consortium.
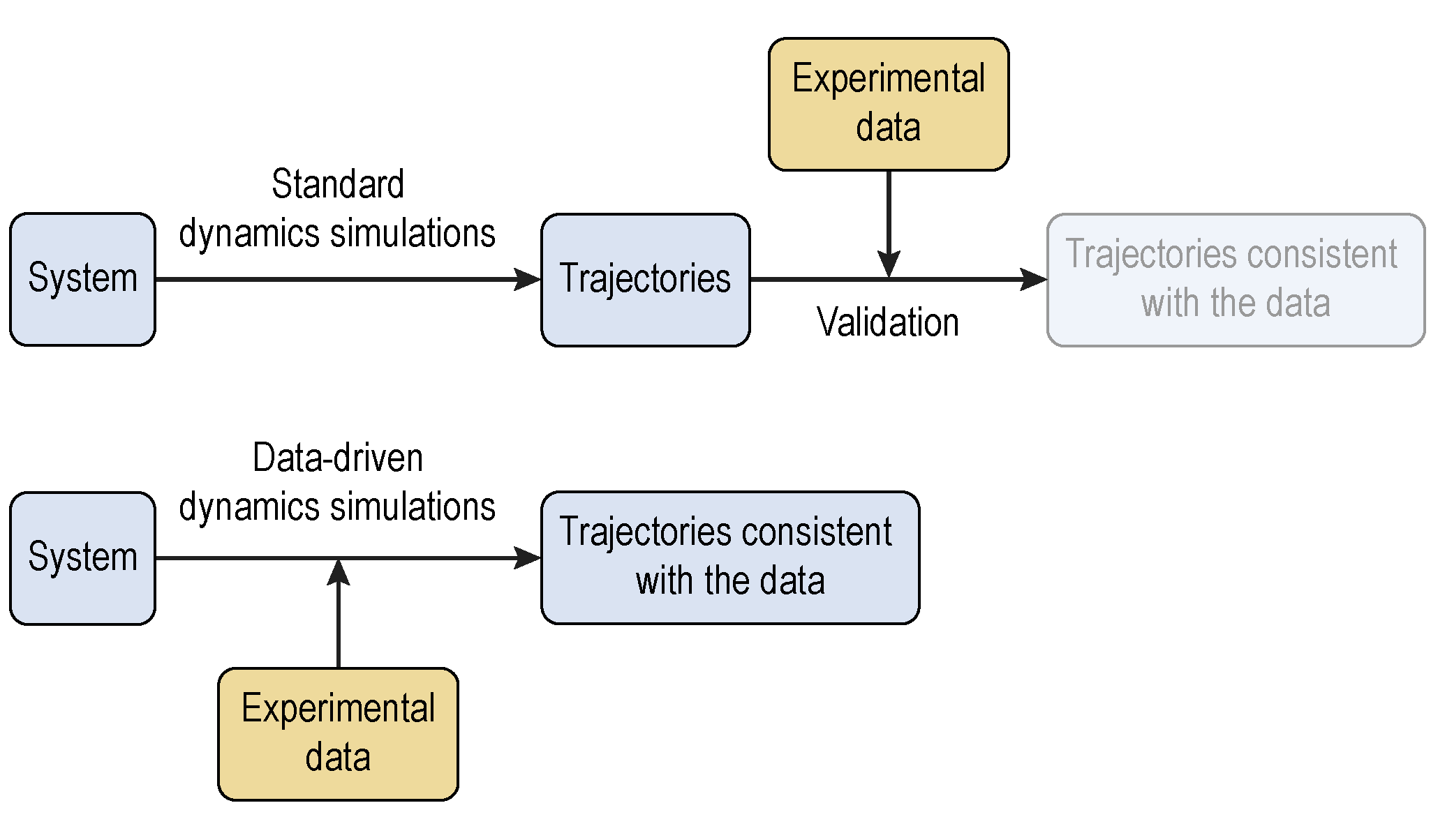
Data-Driven Brownian Dynamics Simulations
Structural models of macromolecular assemblies are increasingly computed via integrative modeling approaches. Despite the importance of studying their dynamic structures, the amount of spatiotemporal models is limited, which are typically constructed based on first physical principles. The difficulty is mainly caused by the high degrees of freedom resulting from the large number of particles and the comparatively long time scales. We develop a data-driven spatiotemporal modeling approach using coarse-grained Brownian dynamics simulations. By construction, the simulated trajectories match experimentally observed properties. The resulting model is therefore more accurate than was done previously, and is expected to be quantitative and predictive. This approach will aid in improving Brownian dynamics simulations and be applicable to model many other dynamic structures.
Biological Image One-Stop Solution (BiOS)
BiOS (Biological image One-stop Solution) provides contextualized one-stop solutions for images in biology and medicine from processing, segmentation, analysis to new discoveries. We have developed auto-segmentation methods to produce organelle masks, post-processing tools to separate individual organelles, and systematic analysis pipelines to identify organelle morphologies and organizations. We have applied these tools on soft X-ray tomograms (SXT) for the fast and accurate generation of insulin vesicle, mitochondrion, nucleus and whole cell masks, and for the characterization of both molecular densities and spatial arrangements of insulin vesicles and mitochondria to reveal the impact of external treatments on subcellular structures. Recently, we are processing through fluorescent movies and cryo-electron tomograms (cryo-ET) to explore velocity distributions, packing properties, and neighborhoods of different organelles under different physiological conditions. Our platform is expected to extend to other image types including MRI, CT, PET-CT, etc.. For more details, please visit BiOS.
Using Cryo-Electron Tomography towards Cell Mapping
F-actin plays an important role in the process of insulin release in pancreatic β-cells. It facilitates secretory granule transportation, docking, and fusion to the plasma membrane. However, the architecture, dynamics, function and relationship with other components still need to be established. Here, we investigate the structure and dynamics of cortical F-actin by using immunofluorescence microscopy, live-cell imaging and cryo-electron tomography under different pseudo-physiological conditions at different time points, in collaboration with Wolfgang Baumeister, Raymond Stevens, and Guisheng Zhong’s labs. We first measure the G-/F-actin ratio using western blot. Via fluorescence microscope, we monitor F-actin dynamics including its polymerization / depolymerization, diffusion, remodeling, as well as F-actin interactions with insulin granules and microtubules. Moreover, we reconstruct actin architecture using cryo-ET. Mapping the structural organization of actin filaments, their spatial relationship with other cellular components, and F-actin dynamics and remodeling process during insulin secretion will provide insights into the biochemical characteristics, physical features, fine structures, and principles of its biological functions. In addition, these data are expected to contribute to the design of the pancreatic β-cell model.